BT Question Set P1-T2-20-24-3: AIC and BIC
20.24.3. Eric is a commodity analyst who fit four different candidate AR(p) models to a series of oil prices. For each of the candidate models, he then retrieved the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). The results are displayed below.
.. AIC and BIC values (table below) goes here …
As Eric interprets the results of these criteria (i.e., the AIC and BIC values), which of the following statements is TRUE?
- Both criteria (AIC and BIC) favor the AR(1) model
- Both criteria (AIC and BIC) favor the AR(2) model
- As both criteria are insignificant, any AR(p) model is a bad fit
- The criteria disagree and therefore the AIC’s preference should prevail because it prefers fewer lags
library(tidyverse)
library(scales)
library(gridExtra)
library(forecast)
library(tseries)
library(fpp2)
library(gt)
library(astsa)
# 175
set.seed(175)
ar2 <- arima.sim(model=list(order=c(2,0,0),ar = c(1.5,-.75)),n = 500)
ma2 <- arima.sim(model=list(order=c(0,0,2),ma = c(1.5,-.75)),n = 500)
ar1_fit <- sarima(ar2, p = 1, d = 0, q = 0)## initial value 1.185288
## iter 2 value 0.473121
## iter 3 value 0.473113
## iter 4 value 0.473112
## iter 5 value 0.473108
## iter 6 value 0.473099
## iter 7 value 0.473095
## iter 8 value 0.473094
## iter 9 value 0.473092
## iter 10 value 0.473087
## iter 11 value 0.473080
## iter 12 value 0.473076
## iter 13 value 0.473075
## iter 14 value 0.473075
## iter 15 value 0.473075
## iter 16 value 0.473074
## iter 17 value 0.473073
## iter 18 value 0.473073
## iter 19 value 0.473073
## iter 20 value 0.473073
## iter 21 value 0.473072
## iter 22 value 0.473072
## iter 23 value 0.473072
## iter 24 value 0.473072
## iter 25 value 0.473072
## iter 26 value 0.473072
## iter 27 value 0.473072
## iter 27 value 0.473072
## iter 27 value 0.473072
## final value 0.473072
## converged
## initial value 0.475632
## iter 2 value 0.475627
## iter 3 value 0.475615
## iter 4 value 0.475613
## iter 5 value 0.475597
## iter 6 value 0.475583
## iter 7 value 0.475579
## iter 8 value 0.475574
## iter 9 value 0.475573
## iter 10 value 0.475570
## iter 11 value 0.475569
## iter 12 value 0.475569
## iter 13 value 0.475569
## iter 13 value 0.475569
## iter 13 value 0.475569
## final value 0.475569
## converged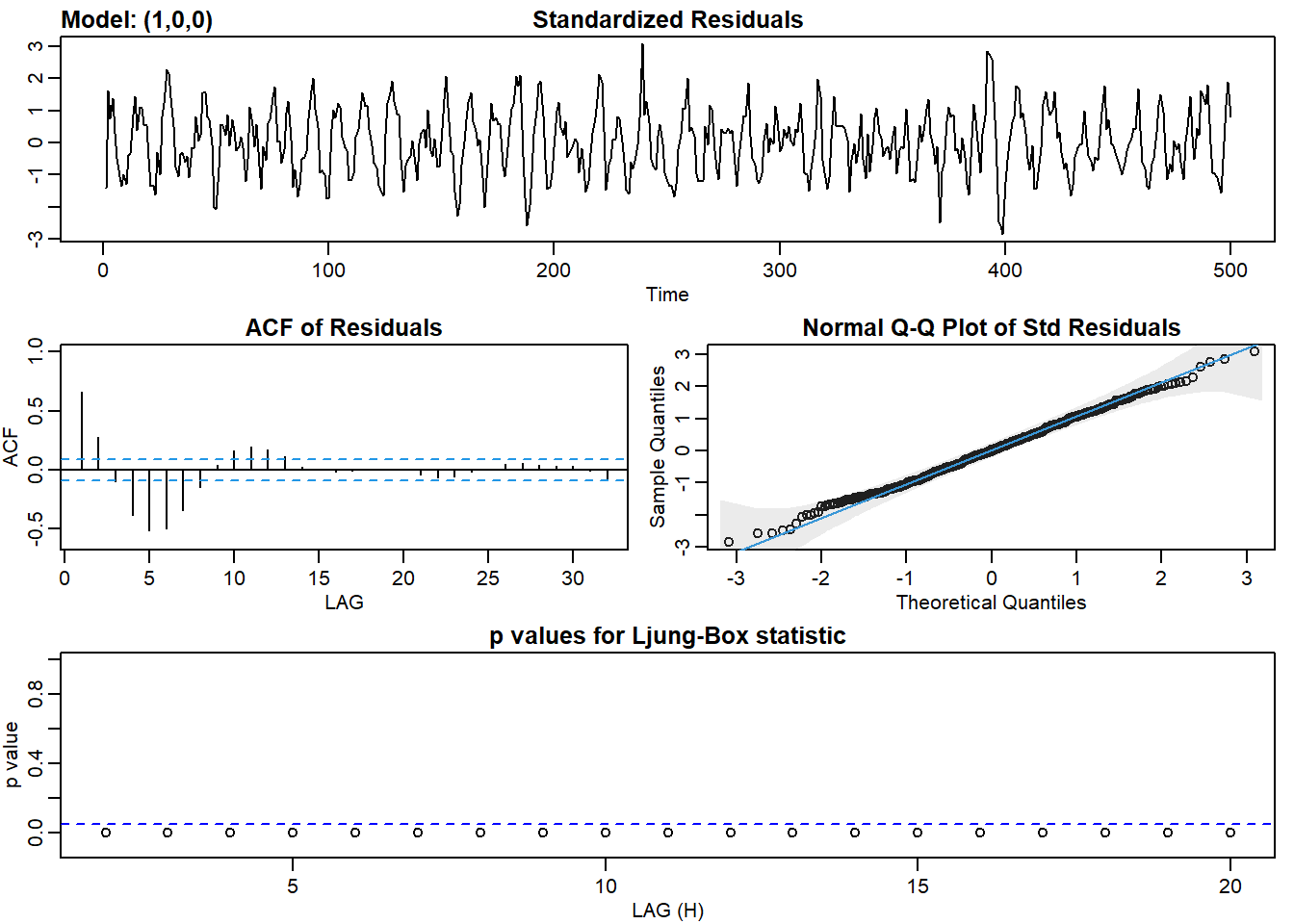
ar2_fit <- sarima(ar2, p = 2, d = 0, q = 0)## initial value 1.186082
## iter 2 value 1.063752
## iter 3 value 0.611728
## iter 4 value 0.360300
## iter 5 value 0.180452
## iter 6 value 0.026222
## iter 7 value 0.023222
## iter 8 value 0.022348
## iter 9 value 0.022310
## iter 10 value 0.022284
## iter 11 value 0.022258
## iter 12 value 0.022258
## iter 13 value 0.022258
## iter 14 value 0.022258
## iter 15 value 0.022258
## iter 16 value 0.022258
## iter 17 value 0.022258
## iter 17 value 0.022258
## iter 17 value 0.022258
## final value 0.022258
## converged
## initial value 0.028108
## iter 2 value 0.028095
## iter 3 value 0.028087
## iter 4 value 0.028086
## iter 5 value 0.028086
## iter 6 value 0.028085
## iter 7 value 0.028085
## iter 8 value 0.028085
## iter 8 value 0.028085
## final value 0.028085
## converged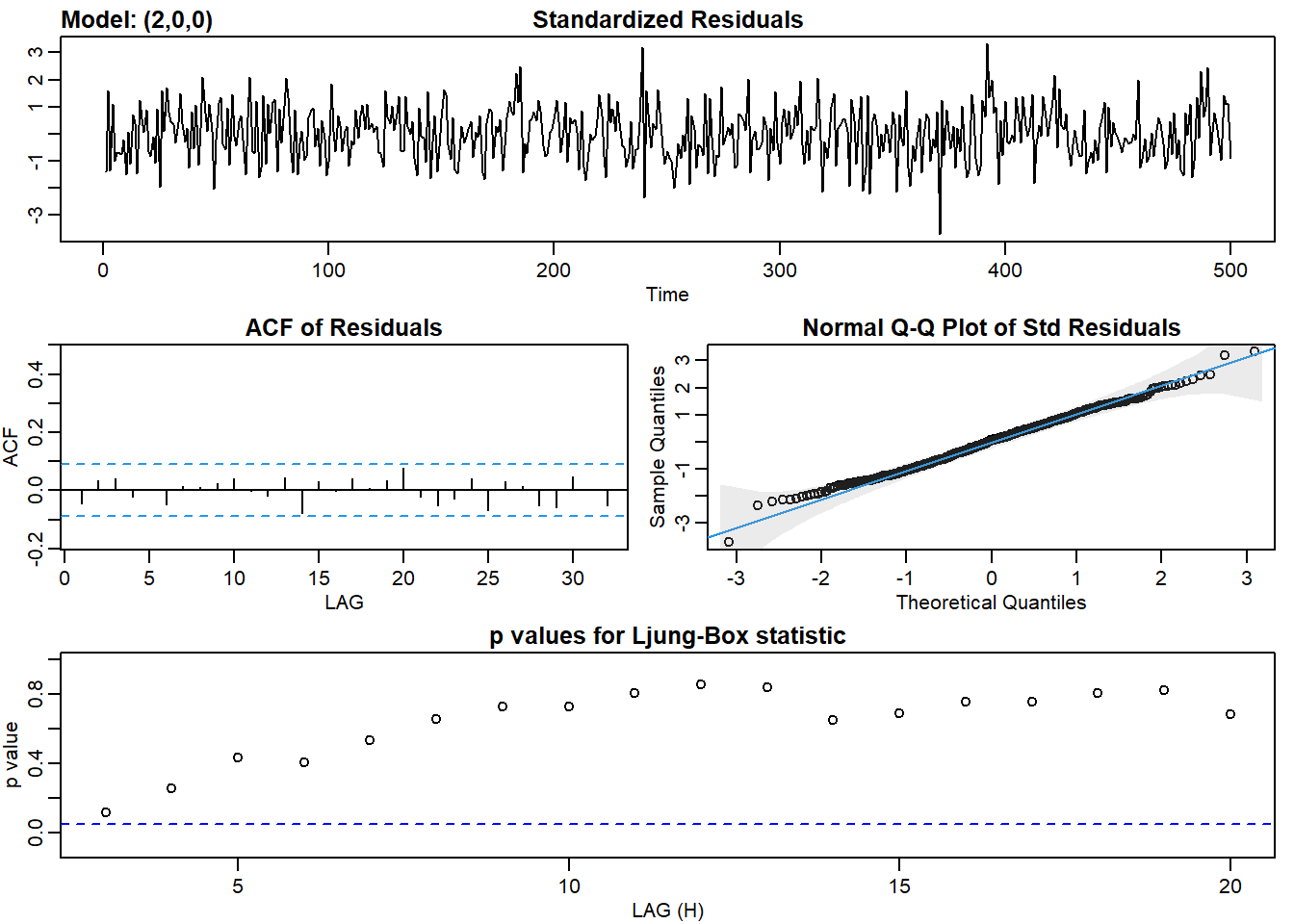
ar3_fit <- sarima(ar2, p = 3, d = 0, q = 0)## initial value 1.187086
## iter 2 value 0.966297
## iter 3 value 0.720122
## iter 4 value 0.651535
## iter 5 value 0.270365
## iter 6 value 0.157749
## iter 7 value 0.106223
## iter 8 value 0.068752
## iter 9 value 0.027592
## iter 10 value 0.020453
## iter 11 value 0.020449
## iter 12 value 0.020445
## iter 13 value 0.020430
## iter 14 value 0.020418
## iter 15 value 0.020418
## iter 16 value 0.020418
## iter 17 value 0.020418
## iter 18 value 0.020418
## iter 19 value 0.020418
## iter 20 value 0.020418
## iter 20 value 0.020418
## iter 20 value 0.020418
## final value 0.020418
## converged
## initial value 0.027093
## iter 2 value 0.027090
## iter 3 value 0.027084
## iter 4 value 0.027079
## iter 5 value 0.027079
## iter 6 value 0.027078
## iter 7 value 0.027078
## iter 8 value 0.027078
## iter 8 value 0.027078
## iter 8 value 0.027078
## final value 0.027078
## converged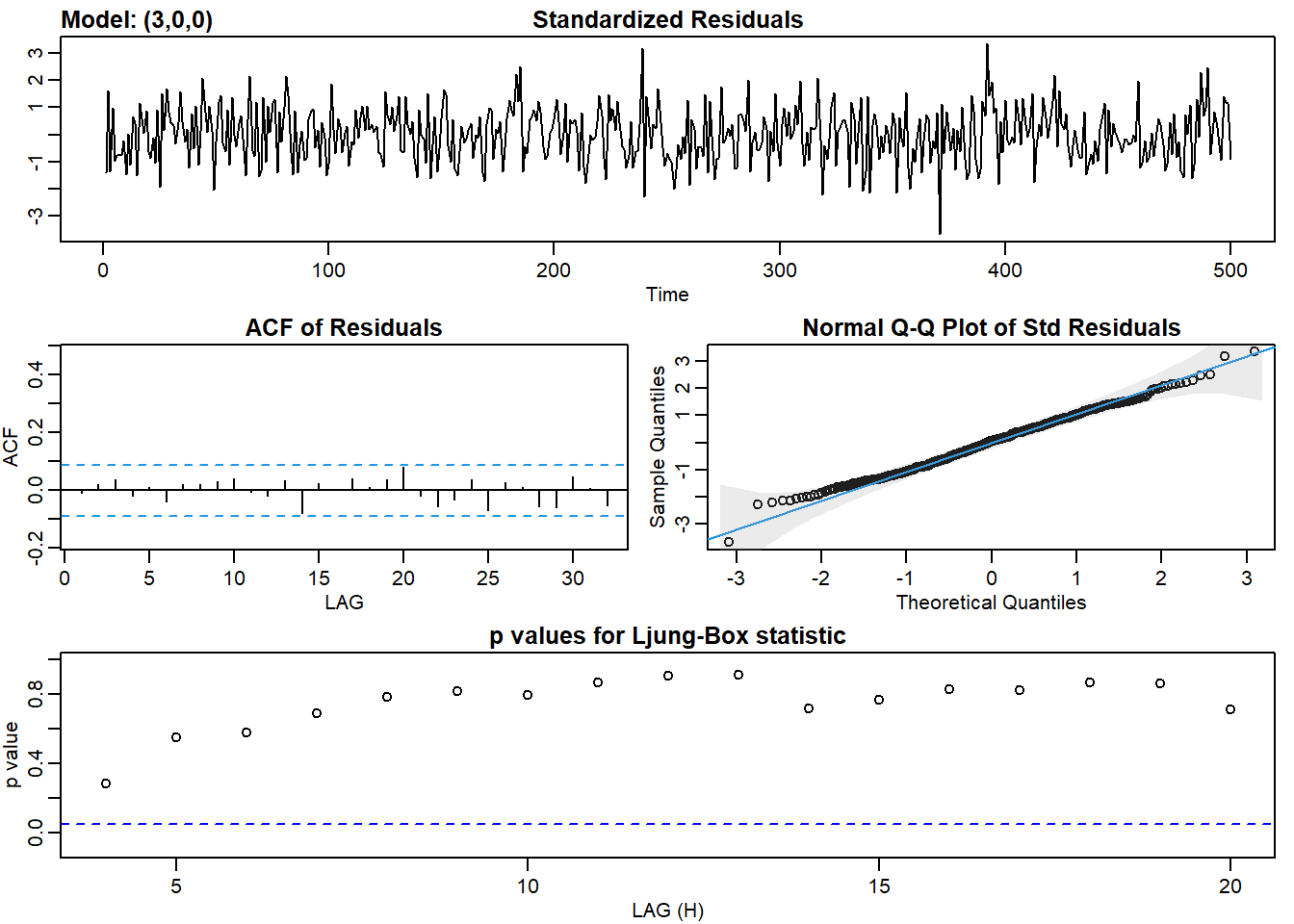
ar4_fit <- sarima(ar2, p = 4, d = 0, q = 0)## initial value 1.187677
## iter 2 value 1.182760
## iter 3 value 0.389837
## iter 4 value 0.312734
## iter 5 value 0.257361
## iter 6 value 0.134886
## iter 7 value 0.042493
## iter 8 value 0.035479
## iter 9 value 0.028843
## iter 10 value 0.020356
## iter 11 value 0.020308
## iter 12 value 0.020305
## iter 13 value 0.020305
## iter 14 value 0.020305
## iter 15 value 0.020304
## iter 16 value 0.020304
## iter 17 value 0.020304
## iter 18 value 0.020304
## iter 19 value 0.020304
## iter 20 value 0.020304
## iter 21 value 0.020304
## iter 22 value 0.020304
## iter 22 value 0.020304
## iter 22 value 0.020304
## final value 0.020304
## converged
## initial value 0.026957
## iter 2 value 0.026941
## iter 3 value 0.026940
## iter 4 value 0.026938
## iter 5 value 0.026937
## iter 6 value 0.026936
## iter 7 value 0.026936
## iter 8 value 0.026936
## iter 9 value 0.026936
## iter 9 value 0.026936
## iter 9 value 0.026936
## final value 0.026936
## converged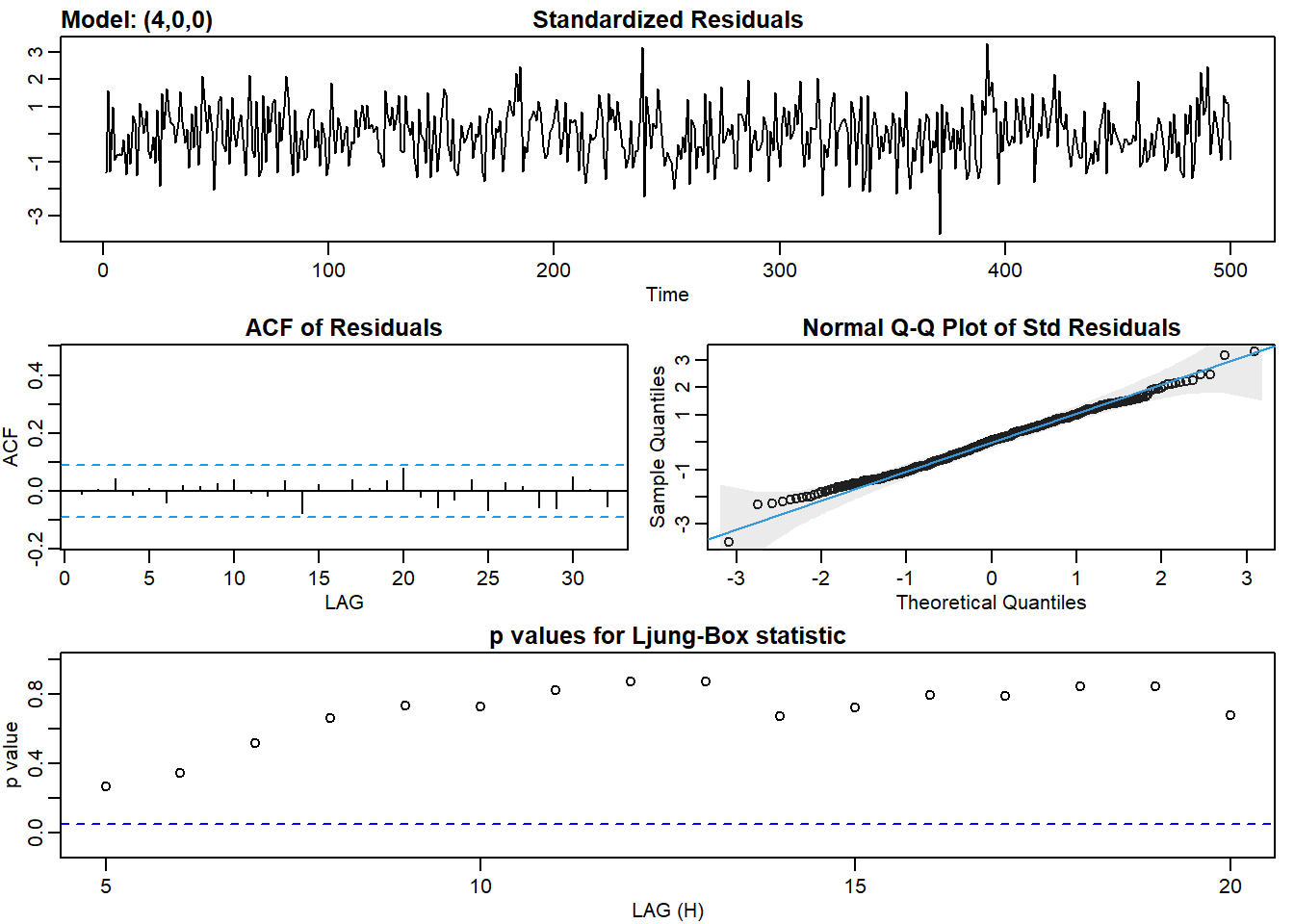
ma1_fit <- sarima(ma2, p = 0, d = 0, q = 1)## initial value 0.620720
## iter 2 value 0.616967
## iter 3 value 0.616503
## iter 4 value 0.616503
## iter 4 value 0.616503
## iter 4 value 0.616503
## final value 0.616503
## converged
## initial value 0.616515
## iter 2 value 0.616515
## iter 2 value 0.616515
## iter 2 value 0.616515
## final value 0.616515
## converged
ma2_fit <- sarima(ma2, p = 0, d = 0, q = 2)## initial value 0.620720
## iter 2 value 0.601563
## iter 3 value 0.601097
## iter 4 value 0.601053
## iter 5 value 0.601053
## iter 5 value 0.601053
## iter 5 value 0.601053
## final value 0.601053
## converged
## initial value 0.601132
## iter 2 value 0.601132
## iter 2 value 0.601132
## iter 2 value 0.601132
## final value 0.601132
## converged
ma3_fit <- sarima(ma2, p = 0, d = 0, q = 3)## initial value 0.620720
## iter 2 value 0.601584
## iter 3 value 0.601080
## iter 4 value 0.600978
## iter 5 value 0.600977
## iter 5 value 0.600977
## iter 5 value 0.600977
## final value 0.600977
## converged
## initial value 0.601058
## iter 2 value 0.601058
## iter 3 value 0.601058
## iter 3 value 0.601058
## iter 3 value 0.601058
## final value 0.601058
## converged
ma4_fit <- sarima(ma2, p = 0, d = 0, q = 4)## initial value 0.620720
## iter 2 value 0.599626
## iter 3 value 0.599377
## iter 4 value 0.599342
## iter 5 value 0.599340
## iter 6 value 0.599340
## iter 7 value 0.599340
## iter 7 value 0.599340
## iter 7 value 0.599340
## final value 0.599340
## converged
## initial value 0.599424
## iter 2 value 0.599424
## iter 3 value 0.599424
## iter 3 value 0.599424
## iter 3 value 0.599424
## final value 0.599424
## converged
print("AIC")## [1] "AIC"ar1_fit$AIC## [1] 3.801016ar2_fit$AIC## [1] 2.910047ar3_fit$AIC## [1] 2.912034ar4_fit$AIC## [1] 2.915749print("BIC")## [1] "BIC"ar1_fit$BIC## [1] 3.826304ar2_fit$BIC## [1] 2.943764ar3_fit$BIC## [1] 2.95418ar4_fit$BIC## [1] 2.966324AIC_ar2 <- c(ar1_fit$AIC, ar2_fit$AIC, ar3_fit$AIC, ar4_fit$AIC)
BIC_ar2 <- c(ar1_fit$BIC, ar2_fit$BIC, ar3_fit$BIC, ar4_fit$BIC)
AIC_BIC_ar2 <- cbind(AIC_ar2, BIC_ar2) %>% data.frame()
models <- c("AR(1)", "AR(2)", "AR(3)", "AR(4)")
models_AIC_BIC <- cbind(models, AIC_BIC_ar2)
models_AIC_BIC_gt <- gt(models_AIC_BIC)
models_AIC_BIC_gt <-
models_AIC_BIC_gt %>%
tab_options(
table.font.size = 14
) %>% tab_style(
style = cell_text(weight = "bold"),
locations = cells_body()
# ) %>% tab_style(
# style = cell_text(color = "cadetblue"),
# locations = cells_column_labels(
# columns = vars(model, 'h (lags)', statistic, 'p-value')
# )
) %>% tab_header(
title = md("**AIC and BIC values**"),
subtitle = "Four candidate models: AR(1) ... AR(4)"
) %>% fmt_number(
columns = vars(AIC_ar2, BIC_ar2),
decimals = 5
) %>% tab_source_note(
source_note = md("AIC = Akaike information criterion")
) %>% tab_source_note(
source_note = md("BIC = Bayesian information criterion")
# ) %>% cols_width(
# vars(model) ~ px(70),
# vars(statistic, 'p-value') ~ px(90)
) %>% cols_label (
models = md("**model**"),
AIC_ar2 = md("**AIC**"),
BIC_ar2 = md("**BIC**")
) %>% tab_options(
heading.title.font.size = 16,
heading.subtitle.font.size = 14
)
models_AIC_BIC_gt| AIC and BIC values | ||
|---|---|---|
| Four candidate models: AR(1) ... AR(4) | ||
| model | AIC | BIC |
| AR(1) | 3.80102 | 3.82630 |
| AR(2) | 2.91005 | 2.94376 |
| AR(3) | 2.91203 | 2.95418 |
| AR(4) | 2.91575 | 2.96632 |
| AIC = Akaike information criterion | ||
| BIC = Bayesian information criterion | ||